 Health & Physiology
Health & Physiology
Keeping CRISPR under control: how bacteria fight viruses without harming themselves
Bacteria are in a constant struggle with the viruses that infect them. While we often think of bacteria as agents of infection, bacteria are in turn infected by viruses, called phage. The phage that infect bacteria and archaea are the most abundant class of organism on earth, with an estimated 10 phage for every bacterial cell.
Bacteria are in a constant struggle with the viruses that infect them. While we often think of bacteria as agents of infection, bacteria are in turn infected by viruses, called phage. The phage that infect bacteria and archaea are the most abundant class of organism on earth, with an estimated 10 phage for every bacterial cell. With more than 1030 bacteria and archaea on earth, this means that phage outnumber stars in the observable universe a billion to one.
Bacteria have evolved a variety of strategies to fight off viral infections. One such strategy is the CRISPR-Cas adaptive immune system (1). CRISPR immunity functions largely at the genetic level. All bacteria and most viruses have genomes composed of DNA, a long molecule composed of a sequence of chemical "letters" that encode all the information required for the bacteria or virus to survive and reproduce. When a virus infects a bacterial cell, it releases DNA directly into the cell. Once it is inside the cell the viral DNA directs the production of many new viruses and ultimately results in the destruction of the bacterium and the release of new viruses that can go on to infect other cells. Components of the CRISPR system can capture a piece of this viral DNA and insert it into their own DNA genome at a unique site called the CRISPR locus. The CRISPR locus acts as a log book of previous infections, with snippets of viral DNA recorded like mugshots. If a virus with a sequence matching a sequence in the CRISPR locus ever reinfects a cell, it can be quickly recognized and destroyed. This system allows the bacteria to adapt to phages that try to infect the cells, giving them a better chance of protecting themselves if the same type of phage infects again, much the same way that a person who gets the flu will develop antibodies that make them resistant to reinfection by the same strain.
The components responsible for capturing pieces of viral DNA are a pair of proteins called Cas1 and Cas2 that work together as a sort of cellular machine. Cas1-Cas2 have the unusual ability to insert these short fragments of DNA precisely at the CRISPR locus. A bacterial genome is composed of millions of DNA letters, and Cas1-Cas2 have to be able to pick out a single specific location to insert the viral DNA. Phage can only be recognized if their DNA is stored at the CRISPR locus, and if Cas1-Cas2 insert the DNA somewhere else, it risks causing a change in the DNA that will hurt or even kill the bacterium. Because of the consequences of sloppy activity, Cas1-Cas2 have evolved to be highly specific.
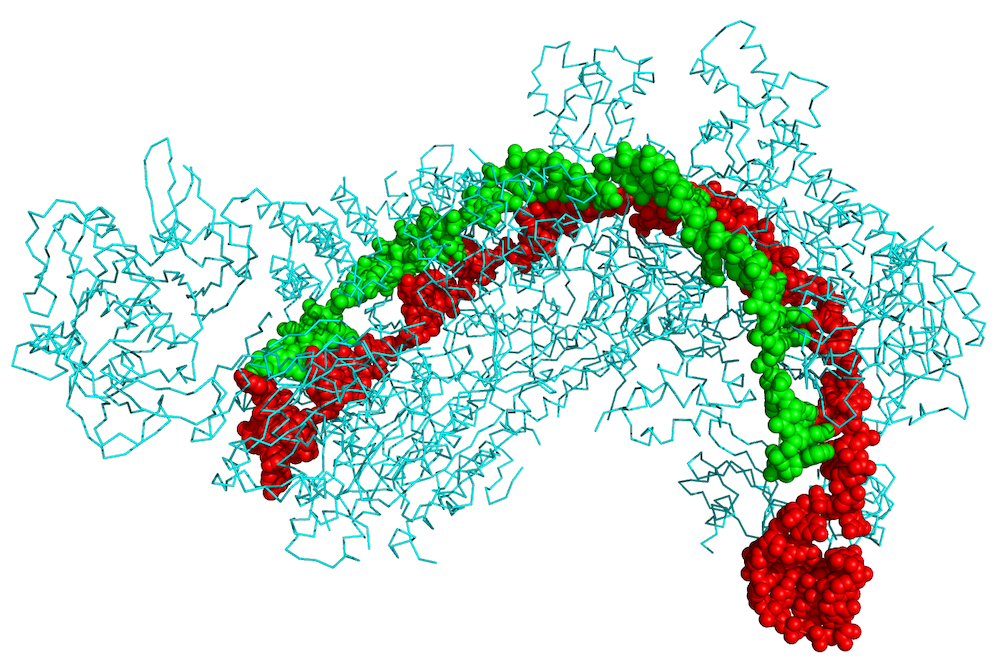
We set out to find how Cas1-Cas2 make sure they only insert DNA at the CRISPR locus. We already knew what sequence of DNA letters at the CRISPR locus were required for insertion, but we didn’t know how Cas1-Cas2 recognize those sequences. We used a pair of techniques called x-ray crystallography and cryo-electron microscopy that allow us to determine the 3D structure of biological molecules in extremely fine detail to see what Cas1-Cas2 look like as they insert new DNA into the CRISPR locus (2). These structures revealed how Cas1-Cas2 recognize the CRISPR locus out of all of the millions of sequences in the bacterial genome.
We found that Cas1-Cas2 identify the CRISPR locus through indirect sequence recognition. While we normally think of the letters of DNA as acting as a code that forms genes and produces the unique characteristics of an organism, these sequences also determine the physical properties of the DNA molecule itself. DNA is a long molecule with a double helix structure, and while it is flexible over long distances, over short distances it acts like a relatively stiff rod. Some sequences will allow that rod to bend in one direction, but not the other, while others might allow the helix itself to partially unwind. Our 3D snapshots of Cas1-Cas2 at the CRISPR locus revealed that the DNA has to bend and unwind just right for the integration reaction to occur. The sequence of the CRISPR locus allows for this twisting to occur, while other sequences are too rigid or bend the wrong direction, preventing integration. In addition, another protein called IHF binds the DNA nearby and forces a 180° bend, bringing another piece of DNA with another recognition sequence into contact with Cas1-Cas2, allowing the proteins to read out both sequences simultaneously. By requiring these unusual DNA distortions, Cas1-Cas2 make sure that they only integrate at the CRISPR locus where the viral DNA can support immunity.
These results help us understand how CRISPR systems work to defend against viruses, but they also have implications for biotechnology. The unique activity of Cas1-Cas2 allows them to act as a molecular recording device, and they’ve even been used to encode a GIF animation into a population of E. coli cells (3). Now that we know how Cas1-Cas2 recognize the CRISPR locus, we can start designing new sequences for them to recognize and expand their use as a tool.
(1) A. V. Wright, J. K. Nuñez, J. A. Doudna, Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell. 164, 29–44 (2016).
(2) A. V. Wright et al., Structures of the CRISPR genome integration complex. Science. 64, eaao0679 (2017).
(3) S. L. Shipman, J. Nivala, J. D. Macklis, G. M. Church, CRISPR–Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature, 1–14 (2017).
Original Article:
A. V. Wright et al., Structures of the CRISPR genome integration complex. Science 357, 1113-1118 (2017)Next read: Surprising Behavior Changes in Genetically Modified Syrian Hamsters by Susan Lee , Kim Huhman , Jack Taylor
Edited by:
Dr. Carlos Javier Rivera-Rivera , Managing Editor
We thought you might like
Driving down malaria
Jul 18, 2017 in Health & Physiology | 4 min read by Andrew Hammond , Xenia Karlsson , Ziyin WangA novel treatment for inherited blinding eye diseases
Feb 1, 2018 in Health & Physiology | 3.5 min read by Tara MooreLego blocks for precise gene editing
Feb 6, 2018 in Health & Physiology | 4 min read by Jared Carlson-Stevermer'Take a deep breath in': a new treatment for congenital lung disease
Dec 20, 2019 in Health & Physiology | 3.5 min read by Vanessa XavierMore from Health & Physiology
Chemotherapy and heart failure
Mar 25, 2024 in Health & Physiology | 2.5 min read by Hector Villarraga , Mariana Garcia ArangoStressing the gut-brain axis
Jan 29, 2024 in Health & Physiology | 3.5 min read by Niklas Blank , Kai Markus Schneider , Christoph ThaissTaurine: a supplement for extending life-span and health
Jan 24, 2024 in Health & Physiology | 3 min read by Vijay Kumar Yadav , Parminder Singh , Kishore GollapalliTake Them Outside: Cold Air Helps Croup Symptoms in Kids
Jan 3, 2024 in Health & Physiology | 3.5 min read by Zoé ValbretHow the immune response to a common virus may target the brain in multiple sclerosis
Dec 20, 2023 in Health & Physiology | 4 min read by Olivia ThomasEditor's picks
Trending now
Popular topics


